Section 6: Extra Information¶
STOP
This section contains information on how to train the classifier for analysing your own data. This will NOT be covered in the workshop.
Train SILVA v138 classifier for 16S/18S rRNA gene marker sequences.
The newest version of the SILVA database (v138) can be trained to classify marker gene sequences originating from the 16S/18S rRNA gene. Reference files and were downloaded from SILVA and imported to get the artefact files. You can download both these files from here.
Reads for the region of interest are first extracted. You will need to input your forward and reverse primer sequences. See QIIME2 documentation for more information.
The classifier is then trained using a naive Bayes algorithm. See QIIME2 documentation for more information.
Requirements and preparation¶
Important
Attendees are required to use their own laptop computers.
At least one week before the workshop, if required, participants should install the software below. This should provide sufficient time for participants to liaise with their own IT support should they encounter any IT problems.
Mode of Delivery
This workshop will be run on a Nectar Instance. For more information click .
It is possible to to run this workshop independently though you will need to install any software needed.
Required Data
-
No additional data needs to be downloaded for this workshop — it is all located on the Nectar Instance. FASTQs are located in the directory and a metadata () file has also been provided.
-
If you wish to analyse the data independently at a later stage, it can be downloaded from here. This zipped folder contains both the FASTQs and associated metadata file.
-
If you are running this tutorial independently, you can also access the classifier that has been trained specifically for this data from here.
Background¶
What is the influence of genotype (intrinsic) and environment (extrinsic) on anemone-associated bacterial communities?
The Players
-
Exaiptasia diaphana — a shallow-water, marine anemone that is often used in research as a model organism for corals. In this experiment, two genotypes (AIMS1 and AIMS4) of E. diaphana were grown in each of two different environments:
- sterile seawater OR
- unfiltered control seawater
-
The anemone-associated bacterial communities or microbiome — these bacteria live on, or within E. diaphana, and likely consist of a combination of commensals, transients, and long-term stable members, and combined with their host, form a mutually beneficial, stable symbiosis.
The Study
The anemone microbiome contributes to the overall health of this complex system and can evolve in tandem with the anemone host. In this data set we are looking at the impact of intrinsic and extrinsic factors on anemone microbiome composition. After three weeks in either sterile or control seawater (environment), anemones were homogenized and DNA was extracted. There are 23 samples in this data set — 5 from each anemone treatment combination (2 genotypes x 2 environments) and 3 DNA extraction blanks as controls. This data is a subset from a larger experiment.
Dungan AM, van Oppen MJH, and Blackall LL (2021) Short-Term Exposure to Sterile Seawater Reduces Bacterial Community Diversity in the Sea Anemone, Exaiptasia diaphana. Front. Mar. Sci. 7:599314. doi:10.3389/fmars.2020.599314 .
QIIME 2 Analysis platform
Attention
The version used in this workshop is qiime2-2021.4. Other versions of QIIME2 may result in minor differences in results.
Quantitative Insights Into Microbial Ecology 2 (QIIME 2) is a next-generation microbiome bioinformatics platform that is extensible, free, open source, and community developed. It allows researchers to:
- Automatically track analyses with decentralised data provenance
- Interactively explore data with beautiful visualisations
- Easily share results without QIIME 2 installed
- Plugin-based system — researchers can add in tools as they wish
Viewing QIIME2 visualisations
As this workshop is being run on a remote Nectar Instance, you will need to and view them in QIIME 2 View (q2view).
Attention
We will be doing this step multiple times throughout this workshop to view visualisation files as they are generated.
Alternatively, if you have QIIME2 installed and are running it on your own computer, you can use to view the results from the command line (e.g. ). opens a browser window with your visualization loaded in it. When you are done, you can close the browser window and press on the keyboard to terminate the command.
Initial Set up on Nectar¶
Byobu-screen
To ensure that commands continue to run should you get disconnected from your Nectar Instance, we’ll .
Reconnecting to a byobu-screen session
If you get disconnected from your Nectar Instance, follow the instructions to resume your session.
Data for this workshop is stored in a central location () on the Nectar file system that we will be using. We will use symbolic links () to point to it. Symbolic links (or symlinks) are just “virtual” files or folders (they only take up a very little space) that point to a physical file or folder located elsewhere in the file system. Sequencing data can be large, and rather than unnecessarily having multiple copies of the data which can quickly take up a lot of space, we will simply point to the files needed in the folder.
Section 5: Exporting data for further analysis in R¶
You need to export your ASV table, taxonomy table, and tree file for analyses in R. Many file formats can be accepted.
Export unrooted tree as format as required for the R package .
Create a BIOM table with taxonomy annotations. A FeatureTable artefact will be exported as a BIOM v2.1.0 formatted file.
Then export BIOM to TSV
Export Taxonomy as TSV
Delete the header lines of the .tsv files
Some packages require your data to be in a consistent order, i.e. the order of your ASVs in the taxonomy table rows to be the same order of ASVs in the columns of your ASV table. It’s recommended to clean up your taxonomy file. You can have blank spots where the level of classification was not completely resolved.
Section 3: Build a phylogenetic tree¶
The next step does the following:
- Perform an alignment on the representative sequences.
- Mask sites in the alignment that are not phylogenetically informative.
- Generate a phylogenetic tree.
- Apply mid-point rooting to the tree.
A phylogenetic tree is necessary for any analyses that incorporates information on the relative relatedness of community members, by incorporating phylogenetic distances between observed organisms in the computation. This would include any beta-diversity analyses and visualisations from a weighted or unweighted Unifrac distance matrix.
Use one thread only (which is the default action) so that identical results can be produced if rerun.
Overview¶
Topic
- Genomics
- Transcriptomics
- Proteomics
- Metabolomics
- Statistics and visualisation
- Structural Modelling
- Basic skills
Skill level
- Beginner
- Intermediate
- Advanced
This workshop is designed for participants with command-line knowledge. You will need to be able to into a remote machine, navigate the directory structure and files from a remote computer to your local computer.
Description
What is the influence of genotype (intrinsic) and environment (extrinsic) on anemone-associated bacterial communities?
Data: Illumina MiSeq v3 paired-end (2 × 300 bp) reads (FASTQ).
Tools: QIIME 2
Pipeline:
Section 1: Importing, cleaning and quality control of the dataSection 2: Taxonomic AnalysisSection 3: Building a phylogenetic treeSection 4: Basic visualisations and statisticsSection 5: Exporting data for further analysis in RSection 6: Extra Information
Section 2: Taxonomic Analysis¶
Assign taxonomy
Here we will classify each identical read or Amplicon Sequence Variant (ASV) to the highest resolution based on a database. Common databases for bacteria datasets are Greengenes, SILVA, Ribosomal Database Project, or Genome Taxonomy Database. See Porter and Hajibabaei, 2020 for a review of different classifiers for metabarcoding research. The classifier chosen is dependent upon:
- Previously published data in a field
- The target region of interest
- The number of reference sequences for your organism in the database and how recently that database was updated.
A classifier has already been trained for you for the V5V6 region of the bacterial 16S rRNA gene using the SILVA database. The next step will take a while to run. The output directory cannot previously exist.
n_jobs = 1 This runs the script using all available cores
Note
The classifier used here is only appropriate for the specific 16S rRNA region that this data represents. You will need to train your own classifier for your own data. For more information about training your own classifier, see .
STOP — Workshop participants only
Due to time limitations in a workshop setting, please do NOT run the command below. You will need to access a pre-computed file that this command generates by running the following: . If you have accidentally run the command below, will terminate it.
Warning
This step often runs out of memory on full datasets. Some options are to change the number of cores you are using (adjust ) or add and try again. The QIIME 2 forum has many threads regarding this issue so always check there was well.
Copy to your local computer and view in QIIME 2 View (q2view).
Visualisation: Taxonomy
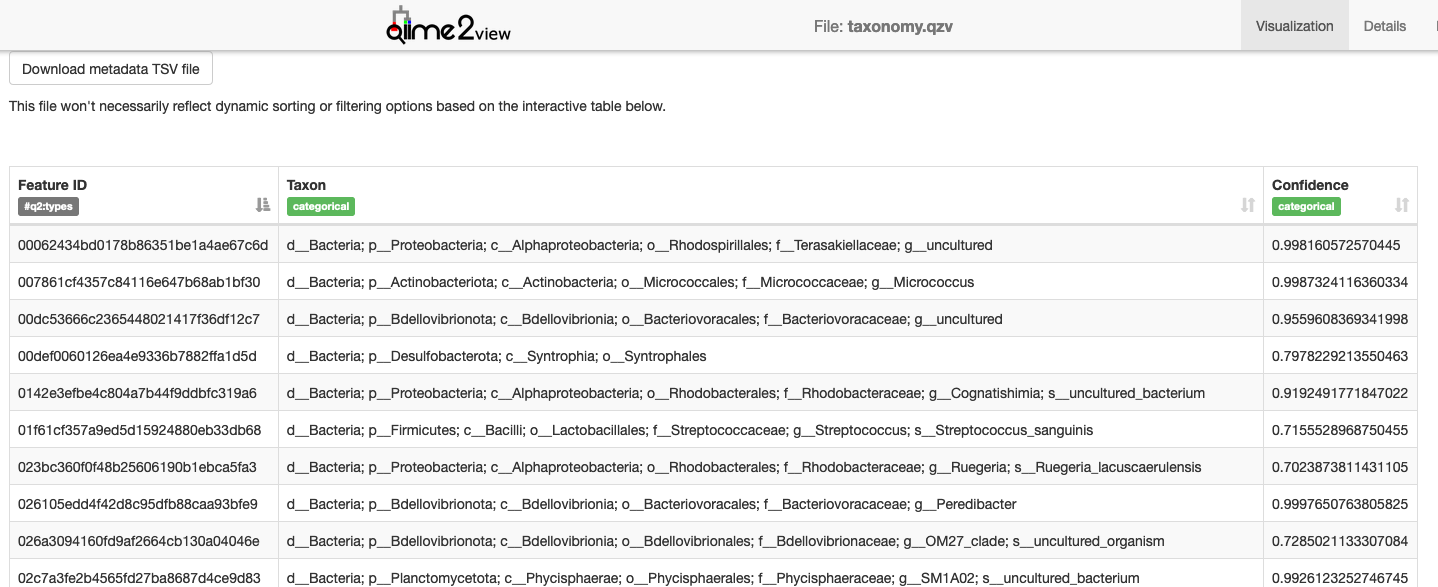
Filtering
Filter out reads classified as mitochondria and chloroplast. Unassigned ASVs are retained. Generate a viewable summary file of the new table to see the effect of filtering.
According to QIIME developer Nicholas Bokulich, low abundance filtering (i.e. removing ASVs containing very few sequences) is not necessary under the ASV model.
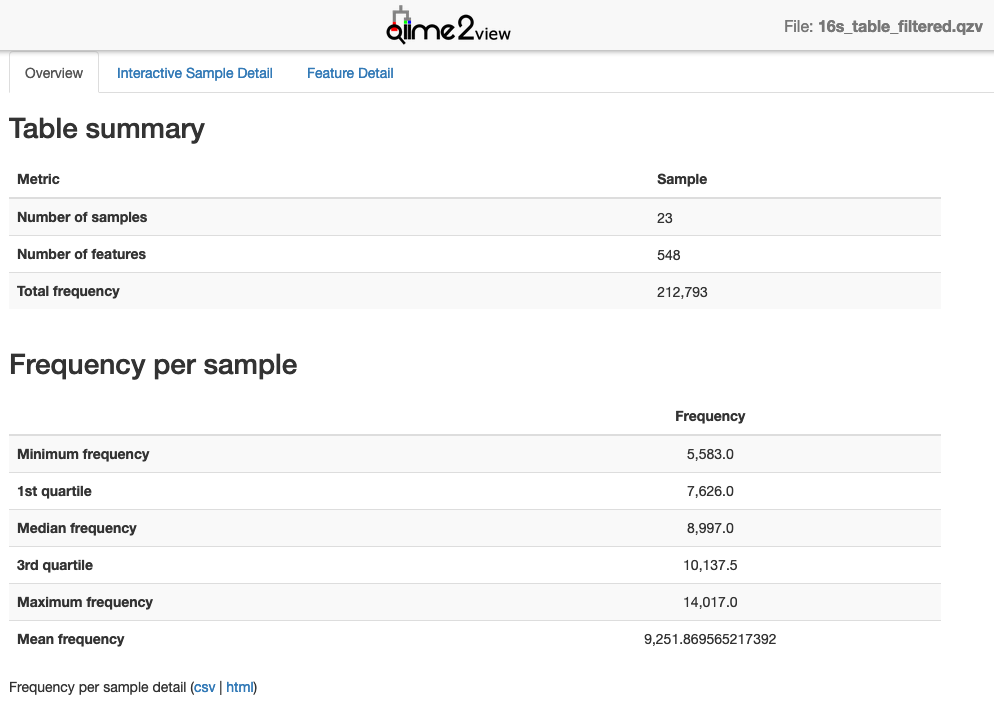
Copy to your local computer and view in QIIME 2 View (q2view).
Visualisation: 16s_table_filtered





























![Volvo prosis offline part & repair [01.2018]](http://dima-sychev.ru/wp-content/uploads/e/5/7/e573819f0dbfa5665baa48f8e74ffbf6.jpeg)